Open code
Reducing spatial autocorrelation in SDM
A simple implementation in R to reduce the negative effect of spatial autocorrelation in distribution models.
Species distribution models (SDM; for review and definition see, e.g., Peterson et al., 2011) are a dominant paradigm to quantify the relationship between environmental dynamics and several manifestations of species biogeography. These statistical approaches pushed an emerging body of research describing the global distribution of species, addressing niche-based questions, supporting biodiversity conservation and ecosystem-based management, as well as inferring the likely anthropogenic pressures leading to population turnover and extinction.
Spatial autocorrelation (SA) is a common challenge while modelling the distribution and abundance of species. This phenomenon, likely present in most ecological datasets, denotes the situation where the values of variables sampled at nearby locations are not independent due to correlation with values at nearby locations (i.e., the value of a predictor variable at a given site can be partially predicted by the values at neighbouring sites).
Accounting for SA has not received much attention in applied SDM studies, however, when present, it may result in poorly specified models and inappropriate spatial inference and prediction. Recent studies proposed to incorporate SA into the actual models while predicting distributions (coined ‘spatial models’; Dormann, 2007), however, this approach does not allow to transfer models to new independent data (e.g., temporal and spatial transferability).
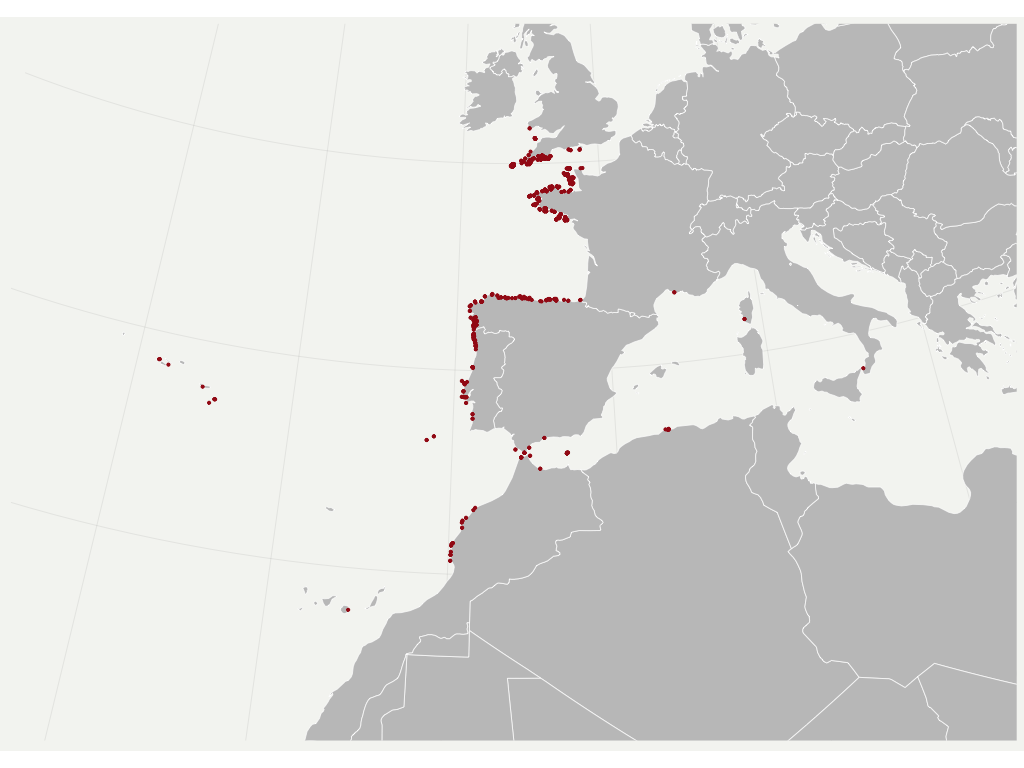
We propose a straightforward approach to reduce the effect of SA in SDM (see also Boavida et al., 2016 for more details). I use a simple example bellow focused on a brown algae species capable of producing marine forests and a set of environmental predictors known to largely explain its distribution.
https://github.com/jorgeassis/spatialAutocorrelation
-
A correlogram is produced to assess the correlation of each variable predictor within a range of geographic distances.
-
For each distance class, a linear model tests the effect of correlation with geographic distance. This finds the minimum non-significant autocorrelated distance.
-
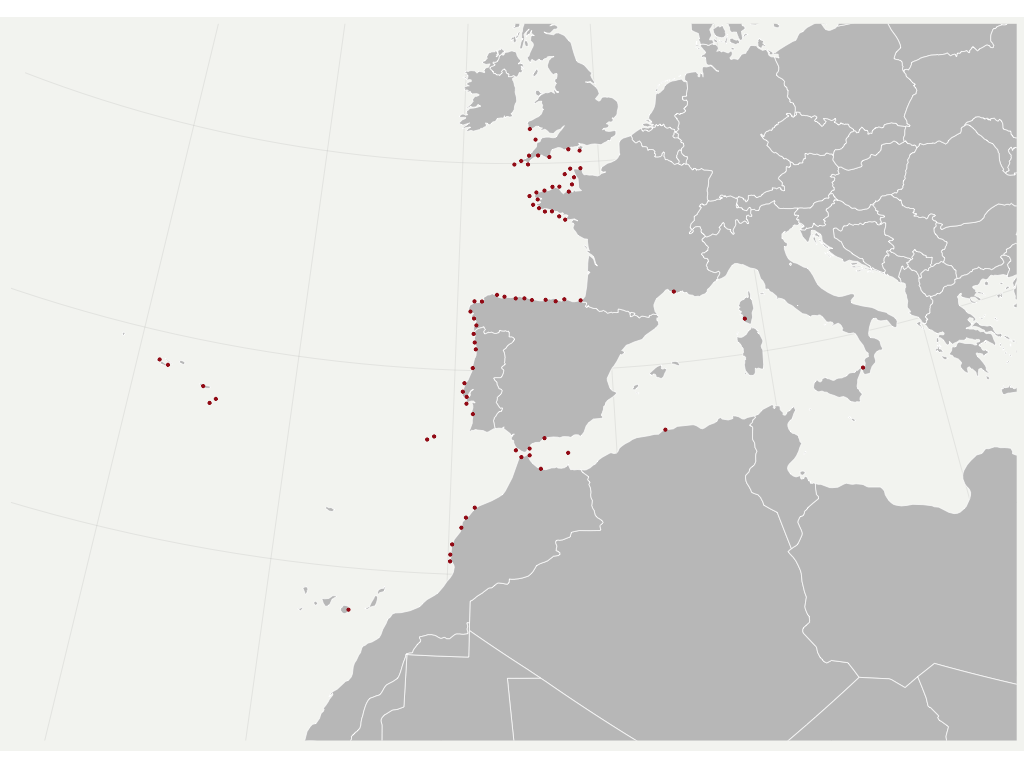
The average of the minimum non-significant distances found per variable is used to prune the occurrence records, by leaving only one record within such distance.
Main reference
Assis, J., Coelho, N. C., Lamy, T., Valero, M., Alberto, F., & Serrão, E. A. (2016). Deep reefs are climatic refugia for genetic diversity of marine forests. Journal of Biogeography, (43), 833–844.
- Featured code
Downloading biodiversity records from iNaturalist
Automatically download biodiversity records from iNaturalist, the most recognised citizen science initiative.
Marine climate layers for ecological modelling
High-resolution marine data layers to model the distribution of species at global scales.